Methods & Protocols
- Home
- Technical Procedures
Technical Literature
In Vivo Link Kit
In Vivo Link Kit for Analysis of Topo I or II Covalent Complex Formation Inside Cells (see ICE Assay Kit TG1020)
Kit Description
TopoGEN has extensive experience with assays for topoisomerase inhibition in vivo. These experiments allow the investigator to ascertain whether a novel agent is active against endogenous topo in a chromosomal setting in nuclei. An important benefit to this analysis is that one can use any tumor cell or tissue in order to establish clinical efficacy of a test drug against a specific tumor cell line. We use essentially the same basic approach for Topo I as for Topo II. The methods are based upon physically separating the topo/DNA adducts from free DNA and using antibodies to measure bound topo I or II. In this analysis, tissue culture cells are treated with a test compound along with negative controls (no drug) and positive controls (with known inhibitors).
The cells are drug treated and rapidly lysed with Sarkosyl which traps some fraction of the endogenous topo on DNA in a covalent cleavage complex. Following detergent lysis, the lysate is diluted to fully dissociate non-covalent DNA/protein complexes. Covalent topo/DNA complexes are resolved on a step CsCl gradient (ionic DNA/protein interactions are also prevented by 5 M CsCl). The gradient resolves DNA, chromatin aggregates, and protein, respectively. Gradients are centrifuged overnight and fractionated. The amount of topo coincident with the DNA peak is a measure of covalent DNA/topo complexes. Topo concentration in the DNA peak is determined by immunoblotting using antibody to topo I or II as probe. In the absence of agents that stabilize the cleavable complex (etoposide, camptothecin) only low levels of topo are found in the DNA peak; this is particularly obvious with topo II since the type II enzyme is not trapped by this method unless an inhibitor is used. In contrast, topo I is trapped to a low extent even in the absence of camptothecin. The ratio of topo at the DNA density and the protein density reflects the relative efficiency of stabilization of the cleavable complexes.
References
Ebert et al., J. Virology 64: 4069-4066, 1990.
Muller and Mehta, Mol. Cell. Biol. 8: 3661-3669, 1988.
Trask and Muller, Proc. Natl. Acad. Sci. 85:1417-1421.
Kit Contents
Camptothecin: supplied lyophilized with Topo I Kits (TG1021)
Etoposide: supplied lyophilized in Topo II Kits (TG1022)
Sarkosyl (20%)
Antibody to Topo I: supplied with Topo I Kits (TG1021)
Antibody to Topo II: 170 kDa form, supplied with Topo II Kits (TG1022)
CsCl Stock Solution
Detailed Instruction Manual
General Procedures
An outline of the method is shown in Fig. 1. Cells to be tested may be a particular cell line, virus infected cell or tumor tissue. Cells are incubated under conditions that favor endogenous topo activity (defined as physiological conditions conducive to cell growth); thus, the endogenous enzyme is engaging the DNA template in a series of breaking and resealing steps. Concurrently, central genetic processes such as DNA replication, transcription and repair are ongoing.
The cells are then rapidly lysed with a detergent (sarkosyl). It is important that lysis be carried out while maintaining the cells at 37 C; if the cells are cooled or manipulated prior to lysis, the cleavage complexes tend to re-ligate and yield negative results (see Trask and Muller, 1988). The next step requires purification of DNA away from free protein; however, organic extractions or proteinase digestions must be avoided. We use a step CsCl gradient for this purpose.
The density steps are designed to resolve DNA from free protein and there is quantitative recovery of both. Covalent complexes containing topo and DNA sediment to the position of DNA. It is known that covalently bound protein can shift the density of DNA in CsCl and the magnitude of the density shift is proportional to the total amount of protein. In vitro, topo I may produce a density shift of DNA under conditions of stoichiometric excess of protein (data not shown); however, in vivo significantly less protein is coupled to DNA. In fact, topoisomerase does not cause any density shift of genomic DNA in this analysis (Fig. 1) for two reasons. First, the number of bound topo mecules per DNA molecule is very low and in these gradients, complexes behave like free DNA (vs. DNA/protein adducts). Second, the gradients are very steep and it is not possible to resolve small density differences anyway. After CsCl centrifugation, the gradients are fractionated and amount of bound and free topo is measured by Western blotting using slot or dot blots and antibodies developed by TopoGEN. The ratio of bound/free is a direct measure of the cleavable complex formation in the particular cell system. We have found that topoisomerases are difficult (if not impossible) to trap as cleavable complexes within cells in the absence of inhibitors; thus, any test compound that results in detection of topo I or II in the DNA peak (Fig. 1) is most likely a topo-active agent. The In Vivo Link-Kit then allows the investigator to evaluate a compound for activity against topo I and II in different tissue settings or with virus infected cells. Finally, it is possible to combine the analysis of topo I and II in the same experiment. In this case, one can evaluate topo I inhibition using the topo I antibody (Cat#2012) or topo II using that antibody (Cat# 2011-1). Some drugs may conceivably act upon both enzymes (Trask and Muller, 1988).
References
1. Ebert et al., J. Virology 64: 4069-4066, 1990
2. Muller and Mehta, Mol. Cell. Biol. 8: 3661-3669, 1988
3. Trask and Muller, Proc. Natl. Acad. Sci. 85:1417-1421
4. Drlica and Franco. 1988. Biochemistry 27:2253-2259.
5. Liu, 1989. Ann. Rev. Biochem. 58:351-375.
6. Osheroff, N. 1989. Pharmacol. Ther. 41:223-241.
7. Wang, 1991. J. Biol. Chem. 266:6659-6662.
8. Wang, J.C. 1985. Annu. Rev. Biochem. 54:665-697.
Methods for Extracting Topoisomerases for Western Blots
Procedure for Making Extracts for Western Blotting. This procedure is designed optimally for HeLa cells; however, other cells lines will work without significant modification. Grow up HeLa cells to near confluence in any convenient culture dish. The following procedure is optimized for a 100 mm petri dish containing about 1‐2 x 107 total cells.
• Decant the medium to waste and swirl/rinse the plate of cells with TD buffer (10‐20 ml per wash/rinse).
• Scrape the cells into 2ml of TD buffer using a rubber policeman.
• Transfer cells into a 15 ml conical centrifuge tube and spin 1000 xg for 10 min at 4 C.
• Discard the supernate and resuspend cells in 2ml of buffer A (per plate)
• Incubate on ice for 10 min
• Spin at 1000xg for 10 min at 4 C to pellet nuclei.
• Discard supernatant, resuspend nuclei in 180ul buffer A (per plate).
OPTIONAL: remove 5‐10 ul of suspension place on microscope slide and evaluate % contamination of whole cells using phase microscope.
• Add 20ul of 10% SDS (sodium dodecyl sulfate, 10% w/v in H2O).
• Sonicate for 15 seconds.
• Add SDS‐PAGE Loading buffer to give a final concentration of 25 mM Tris‐Cl (pH 6.8), 1% SDS, 2.5% 2‐mercaptoethanol,10% glycerol and 0.05% Bromophenol blue.
• Heat samples to 65 C for 5 min.
• Load various amounts of extract on the gel to determine optimal amount of protein that gives best signal:noise ratio on the Western blot. We recommend loading from 0.1 ul to 20 ul for example.
IMPORTANT: If you overload the gels, a lot of extra smeary bands will appear. In this case you should load less protein.
Buffers:
• TD: 100mM NaCl, 20 mM KCl, 0.5mM Na2HPO4, 20 mM TRIS.
• BUFFER A: 100mM NaCl, 50mM KCL 0.1mM EDTA, 20 mM Tris‐Cl, pH 7.5, 0.1mM
• PMSF, 10% Glycerol, 0.2% NP‐40, 0.1% Triton‐X 100
COMPLICATIONS AND IMPORTANT CONSIDERATIONS:
• Fresh extracts work best. Topoisomerase I is especially prone to degradation and you may get subbands that are undesirable as a result.
• Be careful not to overload the gels with excessive amounts of protein! Your gels will look very ugly and many non‐specific bands may come up.
• Topo II is cell cycle regulated. If your cultures are very old or G1 or G0 arrested, your signal may be very weak.
• Topo I can often be present as proteoloytic fragments…protease inhibitors besides PMSF may help (and asnoted, use fresh extracts).
Note that TopoGEN has several different antibodies (rabbit, mouse, human) so it is especially important that you use the appropriate secondary antibody. We find that 125 I‐Protein A works best however, ECL and alkaline phosphatase based detections will also work if optimized.
The basic Western blotting method is standard. There are no other special tricks.
Methods for Extracting Topoisomerases for Enzyme Activity
Small Scale Preparation of Topo I and II Extracts from tissue culture cells (optimized for Hela cells)
Perform all operations using an ice bucket. These enzymes inactivate readily and are easily proteolyzed.
1.From 1–2 100 mm petri dishes, scrape up cells into medium.
2.Pellet cells 800 xg for 3 min in the cold.
3.Resuspend Cell pellet in 3–5 ml of ice cold TEMP buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 4 mM MgCl2, 0.5 mM PMSF) and disperse clumps by pipetting up and down.
4. Repeat centrifugation step and resuspend in 3 ml of TEMP and disperse as above.
5.Leave on ice 10 min.
6.Dounce in tight fitting homogenizer 6–8 strokes (check for nuclei by phase microscopy).
7.Pellet nuclei by centrifugation at 1500 xg for 10 min. (cold).
8.Resuspend the nuclear pellet in 1 ml of cold TEMP. Optional: Transfer to an eppendorf microfuge tube. Repeat step 7 spin or pellet in Microfuge (at 4oC) for 2 min.
9.Resuspend nuclear pellet in a small volume (no more than 4 pellet volumes) of TEP (same as TEMP but lacking MgCl2).
10.Add an equal volume of 1M NaCl, vortex, leave on ice for 30–60 min.
11.Spin in ultrcentrifuge at 100,000 xg for 1 hr. (cold). Alternatively, you may be able to spin in the cold in a microfuge at 12–15,000 xg for 15 min. Ultracentrifugation is best, however.
12.The supernatant will contain topo I and II activities. The type I activity can easily be assayed using the Topo I assay kit (see below) and the type II using kDNA and the TopoGEN Topo II assay kit. Usually, 107 to 108 cells extracted in this way should give large amounts of activity in 1 ul of extract; however, one should titrate over a wide range (from 1:100 dilution in TEMP to as much as 4 ul) to ensure that the reactions are not overloaded. Since the extract contains high salt (0.5M) take care not to poison the reaction with excessive amounts of the extract.
High Temperature Unmasking Technique
High Temperature Antigen Unmasking Technique Using Na–citrate Buffer or IHC with Paraffin Sections
1. Cut and mount sections on slides coated with Vectabond (cat.# SP1800, Vector Laboratories) or Apes (3–aminopropyltriethoxysilane, cat# A3648, Sigma Immunochemicals).
2. Deparaffinize sections and rehydrate with Distilled water.
3. Bring 1.6 l of 0.01M sodium citrate (pH 6) to a boil in a Prestige stainless steel pressure cooker, using a hot plate; cover but do not lock lid.
4. Position slides into metal staining racks and lower into cooker ensuring slides are well immersed in buffer. Lock lid. The small valve will rise.
5. When pressure indicator valve (large one) has risen after about 4 min, incubate sections for 1 min.
6. Remove cooker from heat and run under cold water with lid on. When the small valve sinks, open lid and remove slides and placeimmediately into dist. Water. Don’t open lid until the small valve sinks.
7. Wash sections in TBS (pH7.6) for 1 x 5 min).
8. Place sections in 1.5% hydrogen peroxide/methanol for 10 min.
9. Wash sections in dist. Water for 2 x 5 min; then wash sections in TBS buffer for 2 x 5 min.
10. Place sections in normal serum for 20 min.
11. Cover sections with the primary AB (conditions should be optimized for each system or lab).
12. Wash in TBS buffer for 2×5 min.
13. Incubate sections with secondary AB for 30 min.
14. Was in TBS buffer for 2 x 5 min.
15. Incubate slides in ABComplex for 30 min.
16. Wash in TBS buffer for 2 x 5 min.
17. Incubate slides in DAB .
18. Wash in water for 2 x 5 min.
19. Counterstain with hematoxylin (as needed), dehydrate, coverslip mount and view.
Decatenation-Supercoiling Assay using kDNA
A Decatenation-Supercoiling Assay Using Kinetoplast DNA Suitable for Prokaryotic and Eukaryotic Type II Topoisomerases
ABSTRACT
A versatile assay for eukaryotic and prokaryotic type II topoisomerases is described basedupon detection of kinetoplast DNA (kDNA). Because type II topoisomerases can decatenate DNA but only DNA gyrase can negatively supercoil the products, this assay discriminates between type II topoisomerases that are supercoiling proficient (DNA gyrase) and supercoiling deficient (eukaryotic topo II). The decatenation assay is specific for topo II activity even in crude extracts containing excess topo I. The method is suitable for rapid screening of topo II inhibitors and should be ideal for identification of novel type II enzymes that possess supercoiling activities. Electrophoretic separations are possible within a short time frame (<30 minutes) because of the large size difference between catenated DNA and decatenated products.
INTRODUCTION
A single theme is common to all topos regarding their reaction mechanism: These enzymes adjust the topological state of DNA by breaking and resealing DNA strands, resulting in alterations in DNA linking number (3,11,12). There are two classes of topoisomerases; type II enzymes transiently break both strands of DNA in concert, and type I enzymes transiently break one strand at a time. Given that these enzymes are important targets for novel anticancer and antibacterial therapeutics, assays that high differentiate between the enzyme classes are important in drug discovery (1,3,6). For example, in vitro, type II enzymes efficiently resolve catenated DNAs, such as kinetoplast DNA (4). Another important difference exists between eukaryotic topo II and DNA gyrase (a bacterial topoisomerase II). DNA gyrase uses ATP hydrolysis to negatively supercoil DNA whereas eukaryotic topoisomerase II only relaxes supercoiled DNA. Both enzymes share the same property of being proficient in decatenation reactions in vitro; however, only gyrase has the ability to supercoil the decatenated DNA products. We have modified the original assay described by Marini et al. (4) and show that decatention assays can be used to infer whether a given type II topoisomerase is supercoiling proficient (gyrase) or not (eukaryotic topo II). Additionally, the assay is suitable for identification of novel topo II inhibitors. Because the assay is rapid, it can be adapted for high volume screening of potential inhibitors.
MATERIALS AND METHODS
Enzymes and DNAs
Homogeneously purified human topoisomerase I (TG2005H‐RC1) and topoisomerase II (TG2000H) were used for these studies (9,10). Kinetoplast DNA (TG2013) was also used in these experiments. For details on how we certify enzyme activity (units sold) please see Fig. A1 below.
Enzyme Assays
Eukaryotic topoisomerase II was assayed by decatenation of KDNA and monitoring the appearance of a 2.5 kilobase (KB) DNA. Reactions contained 0.1 ug KDNA (final volume of 20ul), 50 mM Tris‐HCl (pH 8), 120 mM KCl, 10 mM MgCl2, 0.5 mM each of dithiothreitol, ATP and 30 ug BSA/ml (topo II reaction buffer TG4040). The reactions were incubated for 15 min at 37o C and terminated with 0.1 vol of stop buffer (5% sarkosyl, 0.025% bromophenol blue, 50% glycerol). One unit of topoisomerase II is defined as the amount of enzyme required to fully decatenate 0.1 ug of KDNA in 15 min at 37o C. Assays designed to detect topoisomerase II inhibitors were carried out in topo II cleavage buffer (30 mM Tris‐HCl, pH 7.6, 3 mM ATP, 15 mM 2‐mercaptoethanol, 8 mM MgCl2, 60 mM NaCl) in a final volume of 20 ul. Reactions were incubated with 4 units of enzyme in the presence or absence of the indicated inhibitor for 30 min at 37o. The reactions were terminated with 2 ul of 10% sodium dodecyl sulfate, followed by proteinase K treatment for 15 min at 37o. After addition of 0.1 vol of loading dye (50% glycerol, 0.025% bromophenol blue) samples were extracted once with an equal volume of chloroform:isoamyl alcohol (24:1). Following a brief centrifugation in a microfuge, the blue upper layer was loaded directly onto an agarose gel. Assays for DNA gyrase were carried out on reconstituted holoenzyme (equal concentrations of A and B subunits were mixed). Reactions (in a final volume of 30 ul) contained 50 mM Tris‐HCl (pH 7.6), 20 mM KCl, 10 mM MgCl2, 2 mM dithiothreitol, 1.5 mM ATP, 5 mM spermidine, 1.5 ug bovine serum albumin and 0.2 ug KDNA (7). The decatenation products were analyzed on 1% agarose gels run either without or with 0.5 ug ethidium bromide/ml as specified. Electrophoretic analysis of KDNA was performed using standard agarose gel electrophoresis units. Separations designed to resolve supercoiled and relaxed DNA monomers (gyrase assays) were performed at 50 volts. Eukaryotic topo II products were separated at 100 volts which allowed rapid resolution of catenated networks from the minicircles. In gels containing ethidium bromide, appearance of the monomer DNA species was conveniently monitored with a hand‐held UV light source; typically, electrophoresis was continued until the bromophennol blue dye front had migrated 50‐75% down the gel. Following electrophoresis, gels lacking ethidium were stained with 0.5 ug ethidium bromide/ml for 20 min. and photographed. Gels containing ethidium bromide were destained in water (20‐30 minutes at room temperature) prior to photography.
RESULTS AND DISCUSSION
Kinetoplast DNA (KDNA), the mitochondrial DNA of Crithidia fasciculata, is a catenated network of DNA rings, the majority of which are 2.5 KB monomers (8). Type II topoisomerase, but not topo I, has the ability to decatenate KDNA and generate the monomer DNA (4); therefore, decatenation is a highly specific assay for topo II. Furthermore, since KDNA networks are large (relative to the monomers) separation is achieved after only a few minutes of electrophoresis. We modified the original method described previously (4) by running gels in the presence of an intercalator (ethidium bromide) to monitor the appearance of monomers with a hand‐held UV light source and to resolve various DNA forms (linear, nicked circular DNA, supercoiled DNA and relaxed DNA monomers).
A typical reaction is outlined in Figure 1. This gel contains ethidium bromide which allows one to clearly resolve nicked (OC monomers) and circular (relaxed and supercoiled). The kDNA networks are too large to enter the gel, however, purified eukaryotic topoisomerase II yields monomeric DNA rings. In contrast, topoisomerase I did not produce any change in the kDNA networks. The monomer DNA products of eukaryotic topo II are composed of two species, open circular or (OC) DNA and covalently closed circular (CCC), relaxed DNA. Catenated KDNA generally contains some level of OC DNA since the nicking is not caused by the purified topo II (data not shown). In the absence of ethidium bromide, the OC and CCC DNAs are resolved and the relaxed DNA species are present as a gaussian distribution of topoisomers. Inclusion of ethidium bromide in the gel system consolidates the topoisomers into a single band. As noted above, the monomers contained OC DNA due to pre‐existing nicks in the KDNA substrate. (It is concievalbe that nicking occurs during the purification of KDNA since ethidium bromide is used in the purification.) Despite the presence of nicks, topoisomerase I cannot decatenate the networks (not shown). In contrast to the eukaryotic topo II reaction products, gyrase produced decatenated CCC DNA monomers that were supercoiled (last lane on right, Figure 1). The decatenation assay can be used to confirm the presence of a typical eukaryotic type II topoisomerase activity in crude extracts. Topo II will produce monomer species that are OC or CCC DNA, but not linear DNAs. Expected KDNA reaction products for different enzymes are described in Table 1. Formation of linear DNA or degradation products might indicate the presence of a nuclease activity (refer to Table 1). Therefore, in crude extracts a potent DNase I contaminant might interfere. Similarly, in crude extracts, a potent topo I activity might reverse gyrase mediated supercoiling of kDNA monomers. For example, we find that topo II relaxation is able to prevail over gyrase mediated supercoiling (not shown). We conclude that crude extracts containing topo I or II relaxation activity, gyrase supercoiling (but not decatenation) could be masked. To avoid this problem one can use extracts from cells that lack topo I activity, such as null mutations in yeast topoisomerase I or rely on specific topoisomerase inhibitors to suppress a potent relaxation activity. The decatenation assay is also suitable for screening and identification of novel topoisomerase II poisons that stabilize cleavable complexes and lead to nicked or fragmented DNA; however, this should be carried out using purified enzyme (crude extracts may give misleading results). The assay is carried out using purified enzyme and a cleavage buffer optimized to detect formation of cleavable complexes. Cleavage buffer promotes the enzyme into a processive mode (5); thus, decatenation activity is a bit less robust in this buffer. As shown in Figure 1 eukaryotic topo II alone will produced nicked and circular (relaxed) DNA (see Table 1). Inclusion of VM‐26 or VP‐16 (topo II poisons) in the reaction resulted in formation of linears and OC DNA since the inhibitors stabilize single strand and double strand cleavage intermediates). These results require the use of proteinase K in order to resolve topo cleavage complexes since it is necessary to remove the covalently trapped protein off the DNA prior to electrophoresis; failure to do so may shift the band position due to the presence of associated protein. KDNA decatenation assays for drug screening are practical provided that appropriate controls are run (markers, extract alone controls, etc.); however, the investigator must be careful to perform additional controls to ensure that the kDNA is not being nicked or cleaved in a topo II independent fashion. For example, it is conceivable that some test compounds or solvents, may nick DNA. Controls will reveal these cleavage products to be artifactual. Formation of linear or nicked monomer DNA due to the presence of a drug such as etoposide or teniposide is immediately obvious by comparison to control reactions lacking drugs (for more information go to Complications).
There are several advantages to this type of assay:
1. It is fast, allowing high throughput of test samples;
2. The results are clear cut and unambiguous;
3. Topo II activity is robust with kDNA, thus, the enzyme goes farther with this type of assay, and;
4. Decatenation assays can also be used to detect inhibitors that block catalytic activity but do not stabilize cleavable complexes.
REFERENCES
Drlica, K., and R.J. Franco. 1988. Inhibitors of DNA topoisomerases. Biochemistry 27:2253‐2259.
Hajduk, S.L., V.A. Klein, and P.T. Englund. 1984. Replication of kinetoplast DNA maxicircles. Cell 36:483‐492.
Liu, L.F. 1989. DNA topoisomerase poisons as antitumor drugs. Ann. Rev. Biochem. 58:351‐375.
Marini, J.C., K.G. Miller, and P.T. Englund. 1980. Decatenation of kinetoplast DNA by topoisomerases. J. Biol. Chem.
255:4976‐4979.
Muller, M.T., J.R. Spitzner, J.A. DiDonato, V.B. Mehta, and K. Tsutsui. 1988. Single‐strand DNA cleaves by eukaryotic
topoisomerase II. Biochemistry 27:8369‐8379.
Osheroff, N. 1989. Biochemical basis for the inhibition of type I and type II topoisomerases with DNA. Pharmacol. Ther. 41:223‐241.
Otter, R., and N.R. Cozzarelli. 1983. Escherichia coli DNA gyrase. Methods Enzymol. 100:171‐180.
Ryan, K.A., T.A. Shapiro, C.A. Rauch, and P.T. Englund. 1988. Replication of kinetoplast DNA in trypanosomes. Annu. Rev. Microbiol. 42:339‐358.
Spitzner, J.R., and M.T. Muller. 1988. A consensus sequence for cleavage by vertebrate DNA topoisomerase II.
Nucleic Acids Res. 16:5533‐5556.
Trask, D.K., and M.T. Muller. 1983. Biochemical characterization of topoisomerase I purified
from avian erythrocytes. Nucleic Acids Res. 11:2779‐2800.
Wang, J.C. 1985. DNA topoisomerases. Annu. Rev. Biochem. 54:665‐697.
Wang, J.C. 1991. DNA topoisomerases: Why so many? J. Biol. Chem. 266:6659‐6662.
IMPORTANT NOTE about TopoGEN’s Activity Determinations:
For QC activity determinations on topo II provided by TopoGEN, the following conditions apply. Looking at a typical topo II prep (above) you can see that the activity is significantly higher than 2 units/ul (unit defination, 1 unit fully decatenate the 0.2 ug kDNA in 30 min at 37o C). The typical preparation, when fresh, may be anywhere from 6 to 32 units/ul; however we certify at LEAST 2units/ul. For this reason, the customer actually obtains higher activity (=more topo II) than he or she pays for. For example, if you purchase 500 units, you will receive 250 ul of enzyme (@2 u/ul). In fact you are really obtaining from 1000 to as much as 4000 units in your order. We designed it this way because topo II is somewhat unstable and gradual activity loss is unavoidable during shipping; therefore, we send you much more activity than you ordered to account for the unavoiadable loss. For this reason, the actual activity (units/ul) often varies from lot to lot (one lot may be at or near 2 units/ul and the next lot may be much greater than 2 units/ul); however, the activity will never be less than 2/ul as we certify.
For Information on Topoisomerase II assay and Drug Screening Kits, please contact TopoGEN
Technical Services at 614‐451‐5810.
Additional reagents and products used in the preceding article are available from TopoGEN as follows:
Human Topoisomerase II, 170 kDa Form
• TG2000H‐1 250 Units
• TG2000H‐2 500 Units
Human Topoisomerase I, 105 kDa Form
• TG2005H‐RC1 500 Units
Kinetoplast DNA
• Catenated DNA TG2013‐1 25 ug
• Catenated DNA TG2013‐2 50 ug
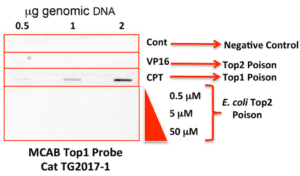
• Linear DNA Marker TG2017‐1 10 ug
• Decatenated Marker TG2020‐1 10 ug
FIGURE A1: TYPICAL QC OF TOPO II LOT OF ENZYME (**INSERT IMAGE**)
FIGURE 1: TOPO II/kDNA
Topo II and DNA gyrase reaction products with kDNA substrate. The data shown below are based upon the following concept: (**INSERT IMAGE**)
Shown below is an idealized view of kDNA reactivity with eukaryotic topo II and prokaryotic DNA gyrase. From left to right:
• kDNA Marker: Most of the kDNA will be high MW catenanes that fail to enter the gel
• Topo II Lest activity: Low enzyme and high enzyme activity levels are shown. At low enzyme activity, some kDNA may remain in the wells; at higher levels, all of the kDNA is released as either OC nicked monomers or relaxed circularized monomer.
• Linear kDNA Marker: Produced by incubating with a restriction enzyme that cuts kDNA monomers once.
• Decatenated kDNA Marker: Shows relative positions of OC nicked and circular monomers
• DNA gyrase products: Since gyrase is a topo II, it will decatenate kDNA; however, it will then supercoil the circular monomers.
Immunoprecipitation with Topo I Monoclonal Antibody
Immunoprecipitation with Topo I Monoclonal Antibody
1. Prepare nuclear protein from HeLa cells (800ug ~ 1mg).
2. Dilute the nuclear protein to approximately 1mg/ml total cell protein with PBS to reduce salt concentration in the buffer.
3. Add the 2ug of the Topo I monoclonalantibody to 1ml of nuclear extract.
4. Gently mix the nuclear extract/antibody mixture for either 2 hours or overnight at 4C on a rocker.
5. Capture the immunocomplex by adding 50ul of protein G sepharose bead slurry (50%) and gently rock for 2 hours.
6. Collect the sepharose beads by pulse centrifugation (5 seconds in the microcentrifuge at 14,000 rpm). Discard the supernatant fraction and wash the beads 3 times with 1ml of ice-‐cold PBS.
7. Resuspend the beads in 30ul of 2 X SDS loading buffer and mix gently.
8. The sepharose beads are boiled for 5 minutes to dissociate the immunocomplexes from the beads. The beads are collected by centrifugation and SDS-‐PAGE is performed with the superna-‐tant fraction.
